Innovative transnational aquatic biodiversity monitoring using high- throughput DNA tools and automated image recognition
Call
Duration
01/03/2024 – 28/02/2027
Total grant
Approx. 2 mil. €
More information
Prof. Dr. Florian LEESE
florian.leese@uni-due.de
Website: www.DNAquaIMG.eu
X: @leeselab
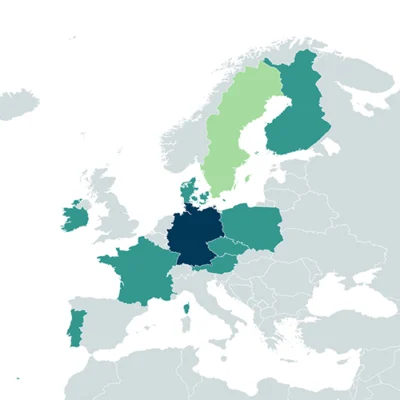
Partners of the project
- Aquatic Ecosystem Research, University of Duisburg-Essen, Essen, Germany
- Program of Environmental information, Finnish Environment Institute (SYKE), Jyväskylä, Finland
- Alpine Centre for research on trophic networks and limnic ecosystems (UMR CARRTEL), French National Institute for Agriculture (INRAE), Food and Agriculture, Food and Environment, Thonon, France
- Department of Invertebrate Zoology and Hydrobiology, Faculty of Biology and Environmental Protection, University of Lodz, Lodz, Poland
- School of Biology & Environmental Science, University College Dublin, Dublin, Ireland
- Faculty of Information Technology, University of Jyväskylä, Jyväskylä, Finland
- Research Centre in Biodiversity and Genetic Resources (CIBIO), BIOPOLIS Association Vairão, Portugal
- Faculty of Science, Department of Geography, Masaryk University, Brno, Czech Republic
- Department of Biology and Ecology, University of Nis, Nis, Serbia
- UNESCO Chair Sustainable Management of Conservation Areas, Carinthia University of Applied Sciences, Villach, Austria
- Phycology Working Group, Faculty of Biology, University of Duisburg- Essen, Essen, Germany
- Department of Mechanical and Production Engineering, University of Aarhus, Aarhus, Denmark
- Department of Water, Atmosphere and Environment, University of Natural Resources and Life Sciences, Vienna, Austria
- Research Group Diatoms, Botanic Garden and Botanical Museum, Berlin, Germany
- Department of Aquatic Sciences and Assessment, Swedish University of Agricultural Sciences, Uppsala, Sweden
- Department of Aquatic Ecology, Faculty of Biology, University of Duisburg-Essen, Essen, Germany
Context
Comprehensive and reliable data are essential to understand biodiversity status and drivers, predict trends, and guide management and restoration in the context of European and international regulations, such as the European Green Deal, the Nature Restoration Law, the EU Biodiversity Strategy and Kunming-Montreal Global Biodiversity Framework. Two emerging approaches hold great promise to improve current biodiversity data generation: DNA-based and automated image-based assessment methods. DNA metabarcoding in particular provides high taxonomic resolution for collected samples and enables the detection of yet undescribed species. In contrast, image-based methods can provide reliable data on species’ abundance, size, structure, and biomass, but can only identify specimens to coarser taxonomic levels (e.g. genus or family). Combined, these complementary approaches offer the potential for greatly enriched biodiversity data, while both approaches can be largely automated to produce FAIR biodiversity data for large sample sizes.
DNAquaIMG proposes to further develop, test, and harmonise DNA-based and automated image-based biodiversity monitoring in rivers by targeting the most typically assessed taxonomic groups in routine freshwater biomonitoring, i.e. invertebrates and diatoms. Building on its results and together with relevant stakeholders, DNAquaIMG will propose a biodiversity monitoring roadmap to fully seize the potential of these new methods into the existing policy context of the EU Water Framework Directive (EG/2000/60, WFD) and to maximise synergies between existing environmental monitoring programmes, jointly improving aquatic biodiversity and ecosystem management.
Main objectives
DNAquaIMG will develop and test an efficient and effective approach to monitor freshwater invertebrates and diatoms with novel DNA and automated image- based biodiversity assessment methods. Together with stakeholders involved in environmental monitoring, the consortium will develop a roadmap for European freshwater biodiversity monitoring using these methods, within the existing and well-established context of the WFD.
Main activities
The performance of the DNA-based and automated image-based methods for freshwater biodiversity monitoring of diatoms and invertebrates will be tested using 150 samples from rivers in eight different countries. To improve processing speed of invertebrate environmental samples, automated procedures, including a robotic arm for sorting, will be developed. To maximise biodiversity data comparability, formal standardisation of DNA-based methods will be initiated within CEN or ISO. Building on these outcomes and together with national and international stakeholders, DNAquaIMG will co-develop a roadmap for these novel methods’ uptake as part of the WFD and other key directives for improved and harmonised biodiversity monitoring in Europe to support better decision-making.