“Monitoring of soil biodiversity: Challenges and lessons learnt from the second year of the Biodiversa+ pilot ‘Soil biodiversity in protected, near-natural forests’”
Published: March 2025 | DOI: 10.5281/zenodo.15261960
Improving the monitoring of the rich ecosystem beneath our feet is one of Biodiversa+’s key priorities. As part of this effort, the pilot project “Soil biodiversity in protected semi-natural forests” was launched in January 2023. The pilot’s objectives include developing feasible experimental design and common protocols, testing eDNA methods for high-resolution data, evaluating the applicability of soil biodiversity-related Essential Biodiversity Variables (EBVs), linking the pilot to international and EU policies, and assessing transnational coordination and governance.
The second-year report of the pilot focuses on methodological challenges, comparing traditional sampling techniques with molecular methods.
Traditional methods
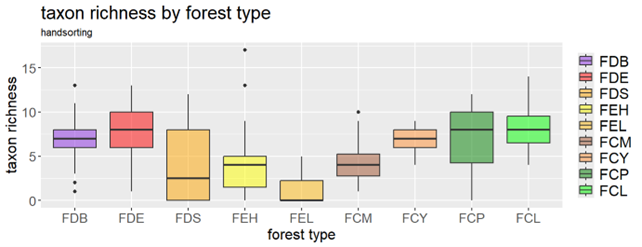
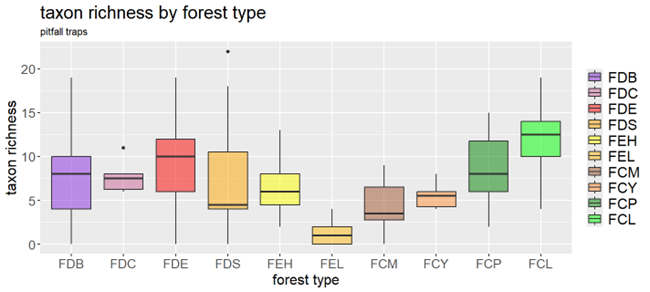
Pitfall traps and hand-sorting of soil cores are well-established methods for monitoring soil macrofauna.
- Pitfall traps are inexpensive, easy to set up, and effective for measuring the activity density of highly mobile invertebrates. They are weather-resistant and not subject to observer bias. However, they require regular maintenance, can be damaged in the field, and may capture non-target species.
- Hand-sorting of soil cores allows for direct collection and counting of soil-dwelling invertebrates. It is also low-cost and requires minimal equipment, but is time-consuming and can introduce observer bias.
While both methods are valuable for detecting macro-invertebrate diversity at the family level, achieving species-level identification typically demands specialised expertise.
Molecular methods
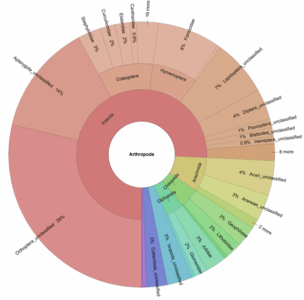
eDNA metabarcoding is a promising and scalable approach for assessing entire soil communities from a single sample. It is faster, less invasive than traditional methods, enables centralised processing, and has the potential to detect a broader range of taxa.
However, several technical challenges remain:
- It requires stringent contamination control and specialised lab equipment.
- The commonly used universal 18S primers (as in LUCAS and SoilBON protocols) have limited taxonomic resolution, which can lead to incomplete or biased results.
- The standard 0.25g of soil used for DNA extraction appears insufficient to capture the full diversity of soil animals.
- Incomplete reference libraries limit taxonomic identification.
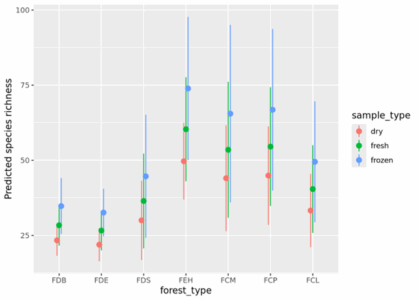
- Drying and freezing soil samples in the lab can reduce alpha diversity. Freezing in the field is preferable, but can be challenging.
- eDNA primarily provides relative abundance data, with PCR amplification bias affecting absolute abundance estimates.
A few figures
Key takeaways
- Traditional methods remain valuable, particularly for macro-invertebrates, but are time-consuming and limited in taxonomic resolution.
- eDNA methods offer enormous potential for scaling up monitoring but need refinement in several areas before they can become standard practice.
- Ongoing work will focus on refining protocols, selecting relevant EBVs, and addressing open questions around sampling design, seasonality, and data use.
By testing and comparing approaches, the pilot is identifying best practices, a key step for developing a robust, harmonised monitoring scheme. A step closer to a future where soil biodiversity can be effectively monitored, understood, and protected at scale!